Methods and Applications offers both beginning and experienced investigators a full range of the powerful tools needed for deciphering how proteins interact to form biological networks as well as for unraveling protein-protein interactions in disease in the search for novel therapeutic targets. Authoritative and highly practical Protein-Protein Interactions.
 Overview Of Protein Protein Interaction Technologies And Alternative Download Scientific Diagram
Overview Of Protein Protein Interaction Technologies And Alternative Download Scientific Diagram
Methods Based on.
Protein protein interaction methods. Methods for Detecting ProteinDNA Interactions. Far-western blotting is a molecular biological method which is based on the technique of western blotting to detect protein-protein interaction in vitro. Computational methods for protein-protein interaction analysis are organized into three categories including cluster-quality-based methods node-affinity-based methods and ensemble.
A high sensitivity means that many of the interactions that occur are detected by the screen. Protein protein interaction 1. The in vitro and in vivo methods like affinity purification Y2H yeast 2 hybrid TAP tandem affinity purification and.
Biophysical methods for the study of protein interactions. A high specificity indicates that most of. Proteins interact with DNA through electrostatic interactions salt bridges dipolar interactions hydrogen bonding H-bonds entropic effects hydrophobic interactions and dispersion forces base stacking.
Characterizing proteinprotein interactions through methods such as co-immunoprecipitation co-IP pull-down assays crosslinking label transfer and farwestern blot analysis is critical to understand protein function and the biology of the cell. In in vitro techniques a given procedure is performed in a controlled environment outside a living organism. Schuck P Zhao H.
Interaction prediction is important as it helps researchers make inferences of the outcomes of PPI. Ashish C Patel Assistant Professor Vet College AAU Anand 2. Protein Domains Interactions only possible due to structural domains within the proteins A protein domain is a conserved part of a given protein sequence and tertiary structure that can evolve function and exist independently of the rest of the protein chain Proteins hold structural domains that allow their interaction with and bind to specific sequences on other proteins.
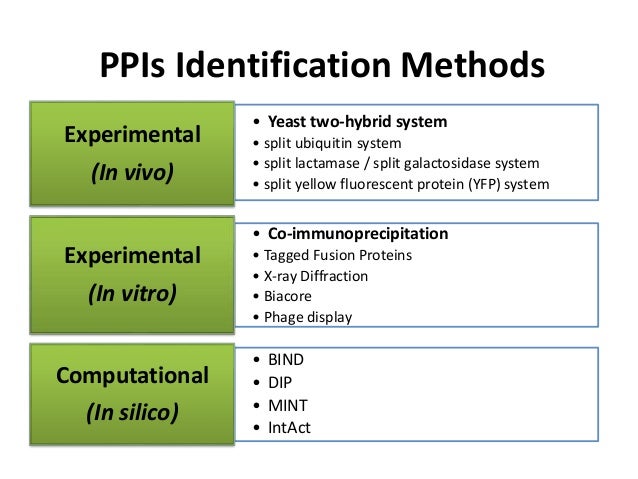
Protein-protein interaction detection methods are categorically classified into three types namely in vitro in vivo and in silico methods. These predictions are done in vivo and various methods can be used to carry out the predictions. Proteins control and mediate many of the biological activities of cells by these interactions.
Split luciferase implementation is a popular method to study protein-protein interactions in planta by using a plant in vivo imaging system to visualize luminescence on transformed leaves. However we still know. The majority of genes and proteins realize resulting phenotype functions as a set of interactions.
BRET Bioluminescence Resonance Energy Transfer was developed to bypass some of FRETs background problems. Protein-protein interactions PPIs control variety of biological phenomena including development cell to cell interactions and metabolic processes The PPIs can be classified into different groups depending upon their functional and structural properties Depending upon their persistence 1 they may be termed as permanent or transient as characterized by their interaction surface 2. Many are physical contacts with molecular associations between chains that occur in a cell or in a living organism in a specific biomolecular context.
There are also empirical methods to detect protein-protein interactions like pulldown assays reviewed in 85 that can be used after identification of candidate genes. Proteinprotein interactions occur when two or more proteins bind together. Each of the approaches has its own strengths and weaknesses especially with regard to the sensitivity and specificity of the method.
Protein-protein interaction plays key role in predicting the protein function of target protein and drug ability of molecules. Authoritative and highly practical Protein-Protein Interactions. Methods and Applications offers both beginning and experienced investigators a full range of the powerful tools needed for deciphering how proteins interact to form biological networks as well as for unraveling protein-protein interactions in disease in the search for novel.
Proteinprotein interaction predictions are methods used to predict the outcome of pairs or groups of protein interactions. These forces contribute in varying degrees to proteins binding in a sequence-specific or. There are many methods to investigate proteinprotein interactions which are the physical contacts of high specificity established between two or more protein molecules involving electrostatic forces and hydrophobic effects.
Firstly because it helps determine the most suitable conditions to use to isolate a particular protein from a mixture of proteins eg pH ionic strength solvent temperature etc and secondly because it may be important to choose conditions which will not adversely affect the molecular structure of the proteins. While usual western blotting uses an antibody to detect a protein of interest far-western blotting uses a non-antibody protein which can bind the protein of interest. Proteinprotein interactions PPIs are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces hydrogen bonding and the hydrophobic effect.